Local Regression
1. Regression with Python
2. Simple Linear Regression
3. Multiple Regression
4. Local Regression
5. Anomaly Detection - K means
6. Anomaly Detection - Outliers
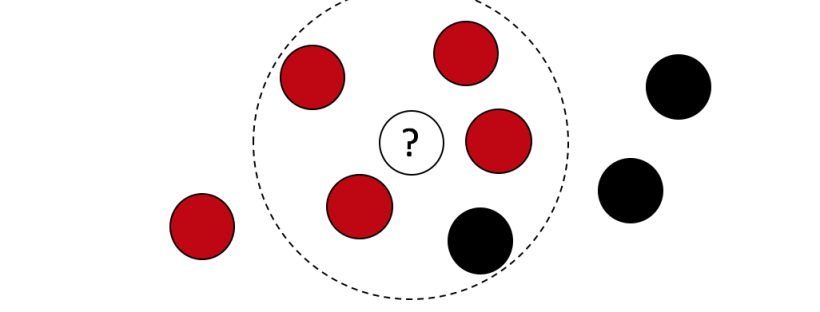
k-nearest Neighbors Regression
In this notebook, you will implement k-nearest neighbors regression. You will:
- Find the k-nearest neighbors of a given query input
- Predict the output for the query input using the k-nearest neighbors
- Choose the best value of k using a validation set

In [None]:
import turicreate as tc
import pandas as pd
import scipy.stats as stats
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inlineIn [None]:
df_sales = tc.SFrame('https://s3.eu-west-3.amazonaws.com/pedrohserrano-datasets/houses-data.csv')In [None]:
df_sales.head()Because the features in this dataset have very different scales (e.g. price is in the hundreds of thousands while the number of bedrooms is in the single digits), it is important to normalize the features
To efficiently compute pairwise distances among data points, we will convert the
SFrame into a 2D Numpy array. First import the numpy library and then copy and
paste get_numpy_data() from the second notebook of Week 2.
In [None]:
def get_numpy_data(data_sframe, features, output):
data_sframe['constant'] = 1
features = ['constant'] + features
features_sframe = data_sframe[features]
feature_matrix = features_sframe.to_numpy()
output_sarray = data_sframe[output]
output_array = output_sarray.to_numpy()
return feature_matrix, output_arrayIn [None]:
(example_features, example_output) = get_numpy_data(df_sales, ['sqft_living'], 'price') # the [] around 'sqft_living' makes it a list
print (example_features[0,:]) # this accesses the first row of the data the ':' indicates 'all columns'
print (example_output[0]) # and the corresponding outputIn [None]:
train_and_validation, test = df_sales.random_split(.8, seed=1) # initial train/test split
train, validation = train_and_validation.random_split(.8, seed=1) # split training set into training and validation setsIn [None]:
feature_list = df_sales.column_names()[3:-1]Using all of the numerical inputs listed in feature_list, transform the
training, test, and validation SFrames into Numpy arrays:
In [None]:
features_train, output_train = get_numpy_data(train, feature_list, 'price')
features_test, output_test = get_numpy_data(test, feature_list, 'price')
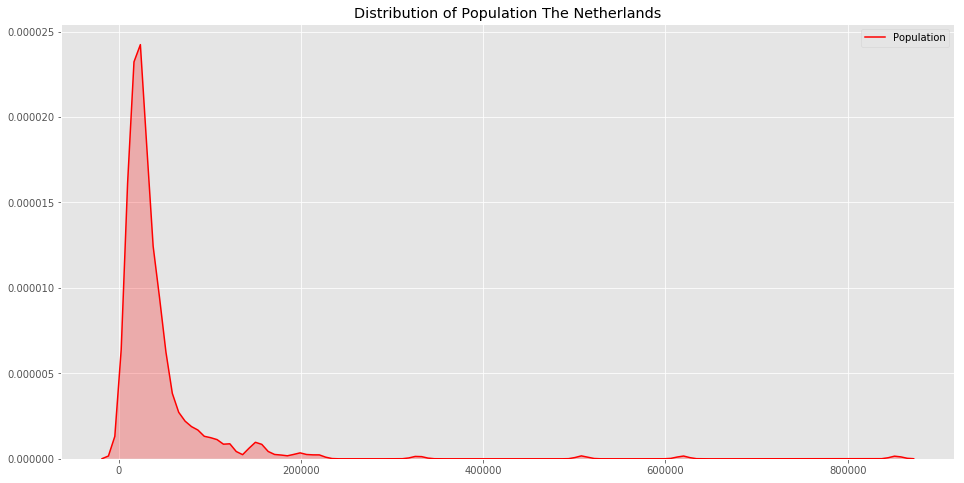
features_valid, output_valid = get_numpy_data(validation, feature_list, 'price')In computing distances, it is crucial to normalize features. Otherwise, for
example, the sqft_living feature (typically on the order of thousands) would
exert a much larger influence on distance than the bedrooms feature (typically
on the order of ones). We divide each column of the training feature matrix by
its 2-norm, so that the transformed column has unit norm.
In [None]:
def normalize_features(feature_matrix):
norms = np.linalg.norm(feature_matrix, axis=0)
normalized_features = feature_matrix / norms
return normalized_features, normsIn [None]:
features_train, norms = normalize_features(features_train) # normalize training set features (columns)
features_train= tc.SFrame(data=pd.DataFrame(features_train))
features_test = features_test / norms # normalize test set by training set norms
features_test = tc.SFrame(data=pd.DataFrame(features_test))
features_valid = features_valid / norms # normalize validation set by training set norms
features_valid = tc.SFrame(data=pd.DataFrame(features_valid))Fitting KNN
Find the (error) way to call the features
In [None]:
knn_model = tc.nearest_neighbors.create(features_train, features = )In [None]:
knn_model.summary()Distance functions
The most critical choice in computing nearest neighbors is the distance function that measures the dissimilarity between any pair of observations.
In [None]:
model = tc.nearest_neighbors.create(features_train, features = ,
distance='manhattan')
model.summary()Distance functions are also exposed in the turicreate.distances module. This allows us not only to specify the distance argument for a nearest neighbors model as a distance function (rather than a string), but also to use that function for any other purpose.
In the following snippet we use a nearest neighbors model to find the closest reference points to the first three rows of our dataset, then confirm the results by computing a couple of the distances manually with the Manhattan distance function.
In [None]:
model = tc.nearest_neighbors.create(features_train, features = ,
distance=tc.distances.manhattan)In [None]:
sf_check = features_train[['0', '1', '2']]
print ("distance check 1:", tc.distances.manhattan(sf_check[2], sf_check[10]))
print ("distance check 2:", tc.distances.manhattan(sf_check[2], sf_check[14]))Search methods Another important choice in model creation is the method. The brute_force method computes the distance between a query point and each of the reference points, with a run time linear in the number of reference points. Creating a model with the ball_tree method takes more time, but leads to much faster queries by partitioning the reference data into successively smaller balls and searching only those that are relatively close to the query. The default method is auto which chooses a reasonable method based on both the feature types and the selected distance function. The method parameter is also specified when the model is created. The third row of the model summary confirms our choice to use the ball tree in the next example.
In [None]:
model = tc.nearest_neighbors.create(features_train, features = ,
method='ball_tree', leaf_size=5)
model.summary()